

 1
2
3
4
5
6
7
8
9
1
2
3
4
5
6
7
8
9

3.2 The descriptions
Starting from the selection of a descriptive model, the program automatically generates a questionnaire [4]. It permits the less informed biologists, as the expert, to acquire personal descriptions and create a case base. An identification name is associated to each observation in order to form a description or a case (Fig. 3).
The description process generates sub-trees of the descriptive model (Fig. 1 and Fig. 3). Therefore, observed descriptions can be directly compared to one another by leafing through page by page: this navigation process is easier than viewing different lists of attribute-value pairs.
In Fig. 3, we illustrate possibilities of IKBS for rendering complete and comprehensive descriptions of a given sample.
Different types of attribute are used: taxonomic ones (e.g. general shape of object colony), numerical intervals (e.g. diameter of apical parts) and multi-nominal values (e.g. section of apical parts). The latter shows variation in objects displaying a set of multiple elements.
The visualization of objects differs graphically according to their status: black if present, black with a cross if absent, dimmed if unknown (see object "hood" at the bottom-right side of Fig. 1 and Fig. 3).
At last, an object can be specialized (e.g. the septa of calices from apical parts, see Fig. 1): the result is a substitution of its name by a more precise one (e.g. primary septa, see Fig. 3) with its associated attributes (inherited or not, see Fig. 2).
It is important for the user to visualize structured descriptions: so doing brings better clarity and comprehensibility to the acquisition phase. This is the most important part of our methodology for acquiring good results of classification and identification.
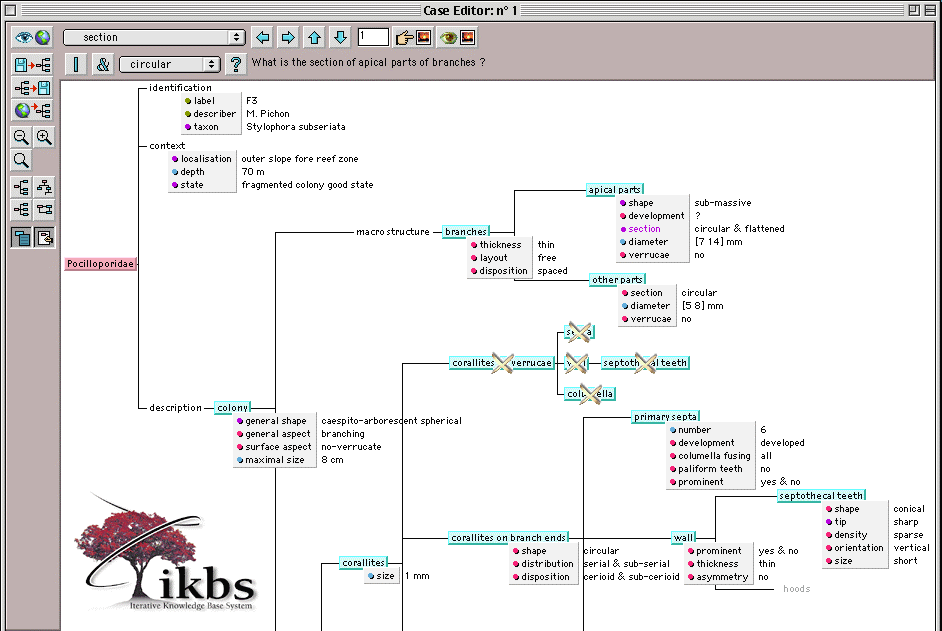
Fig. 3. Part of the description tree of a case of the Family Pocilloporidæ: Stylophora subseriata