

 1
2
3
4
5
6
7
8
9
1
2
3
4
5
6
7
8
9

3. Knowledge acquisition
This part of the methodology is very important to acquire a case base with good quality descriptions, i.e. that are well structured in different dimensions with all the required information, with characters, illustrations and comments that are easily comprehensible for other biologists.
3.1 The descriptive model
The descriptive model represents all the observable characteristics (objects, attributes and values) pertaining to individuals belonging to a particular domain. It is organized in a structured scheme, the name of the domain being at the root of a description tree. Each node of the tree is an object (a component of the individual) defined by a list of attributes with their respective possible values. Designing a descriptive model is essentially an expert task.
For helping them, we have set up logical rules for case description covering: decomposition, viewpoint, iteration, specialization, contextual conditions, etc. [11]. These rules were constructed from the analysis of expert's process of creating monographs of organisms or diseases.
To serve as an example, we present the descriptive model of one of the world's most widespread family of corals: Pocilloporidæ [7] (see Fig. 1). 51 objects and 120 attributes have been defined by the expert. With them, biologists are able to describe 4 genus and 14 species (see attribute called "taxon" in Fig. 1).
There are multiple benefits in such a representation. Viewpoints divide the descriptive model into homogeneous parts, thus giving a frame of reference for describing organisms at a particular level of observation (see objects identification, context, description, macro and micro structure in Fig. 1).
Sub-components introduce modularity into the descriptions making it possible to structure the domain from most general to most particular parts. This object representation of specimens is semantically better than the flat feature-value one: in the former, local descriptions of attributes depend on the existence of parent objects, although in the latter the defined characters are independent of one another. Some of the possibly missing objects are marked with a minus sign (e.g. columella).
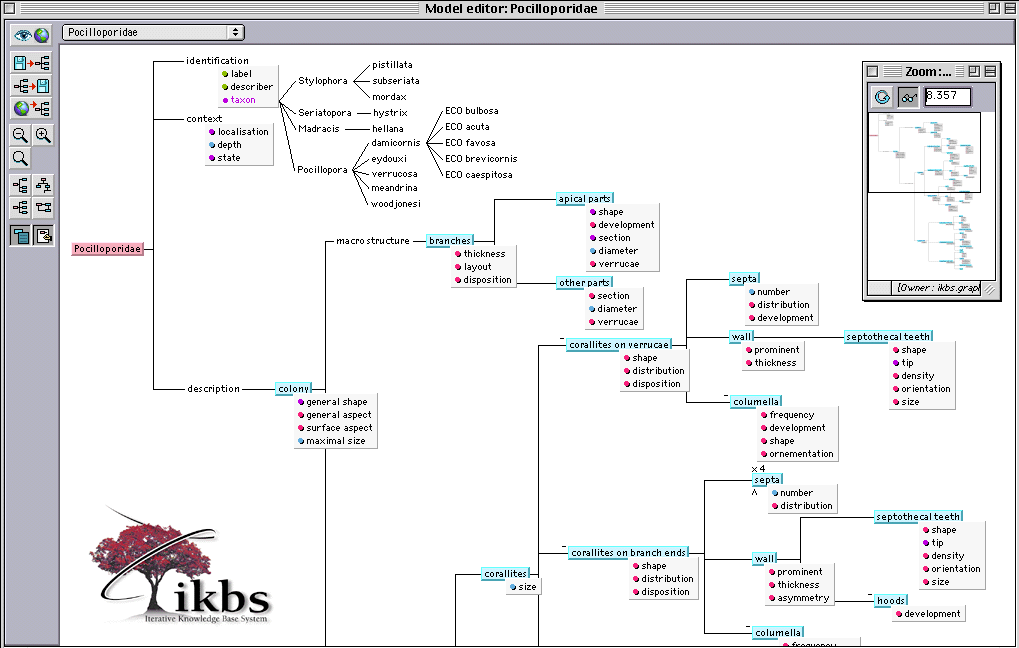
Fig. 1. Part of the descriptive model of the Family Pocilloporidæ
Fig. 1 shows the partitioning dimension of objects (subpart links for disjoint classes). For some of them (i.e. septa), other dimensions such as multi-instantiation (x symbol) and specialization (^ symbol) of objects can be seen. The former enables users to describe several sorts of the same object by descriptive iteration (there are 4 possible instances for septa in Fig. 1) and the latter lets users name each sort with the help of the following classification tree of objects (specialization links in Fig. 2).
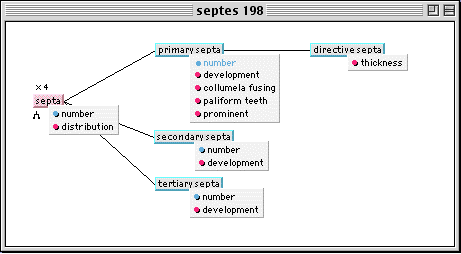
Fig. 2. Classification tree of object "septa"
In fact, one of the roles of the descriptive model is to bring an observation guide for the end-user: the objects are linked together by relations that go from the most general to the most specific (from left to right), making the next description process easier for the non specialist (see below).